The following is a guest post from Karin Strauss from Microsoft Research and Luis Ceze from the University of Washington.
The ability to automate computation has fueled the IT industry for a long time. Since the Babbage machine, our industry has been hard at work at making this process more and more efficient. But is that the only form in which computation can be useful? What about deploying computation in unfriendly environments or volumes where these machines won’t fit?
Wouldn’t it be fantastic if computation can be deployed to a human body and, even more specifically, inside certain types of cells? For example, wouldn’t it be awesome to deploy computation to cells and trigger a specific function only inside cells that are misbehaving, like cancerous cells?
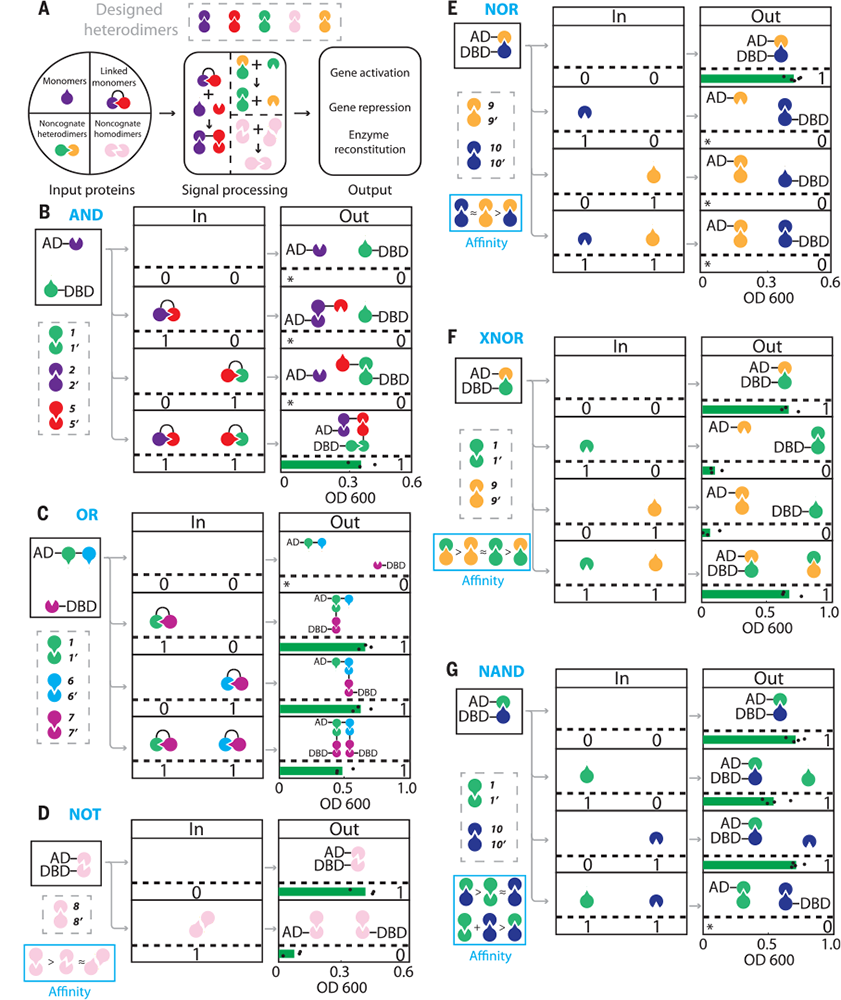
Fig. 2. CIPHR two-input logic gates. From De novo design of protein logic gates. (2020). Science, 368(6486), 78–84. Retrieved from https://science.sciencemag.org/content/368/6486/78. Reprinted with permission from AAAS.
Researchers at University of Washington, Northwestern University, Ohio State University, University of California, San Francisco, and Altius Institute for Biomedical Sciences have taken another step in this direction in a paper recently published in Science (368, 78-84, April 3 2020). The computation form they focus on is protein-based computation, as they are central to natural biological circuits and could be designed or modified to create new therapies and provide other cellular control.
The authors describe four properties that make protein-based circuits more scalable and useful:
- the availability of a large number of orthogonal gates that can be composed into bigger systems without interference;
- modularity, in that the building blocks to be composed should be similar in structure and interchangeable;
- the ability to tune components so their binding affinity with each of multiple partners can be controlled; and
- insensitive to variation of input intensity (when it comes to molecules, concentration) across different inputs – the equivalent to digital inputs despite being represented by analog signals.
They then describe a system that meets all four criteria using proteins they have engineered and attached to natural proteins known to affect cellular behavior of T cells (immune system cells), and demonstrate their utility by using one such circuit to reverse T cell exhaustion, a problem currently faced by solid tumor therapies based on engineered T cell.
In summary, this is one more step in taking computational capabilities to a new domain: the cell. We are excited to see what can happen when engineering and biology come together!









